Research and development
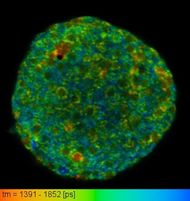
Now I work in Laboratory of Optical and Computational Instrumentation in University of Wisconsin Madison. This lab is mainly a biophotonics lab which focuses on optical and computation technique for imaging. I am primarily involved in FLIM(Fluorescence-lifetime imaging microscopy) imaging technique for producing an image based on the differences in the exponential decay rate of the fluorescence from a fluorescent sample. FLIM is a way of looking into excited state lifetime of a fluorophore. The exponential decay during emission gives us a new layer of information in top of the fluorescence intensity. The lifetime depends the microenvironment of the pixel being excited. FLIM can be thought of as a probe which inspects the microenvironment, pH, metabolic state of cell without any external tagging. One of the prime application of FLIM is looking into shift in metabolism due to cancer. As Warburg hypothesis states, cancer cell depends on Aerobic glycolysis for ATP generation despite the presence of Oxygen. FLIM is one of the best way to study shift in metabolism.
Microglia characterization
Microglia, the resident macrophages of the brain play very important role in neurodegenerative diseases such as Alzheimer's disease, Parkinson's disease, multiple sclerosis etc. Because of microglia’s critical role in nervous system specific immune activities, identification of microglia label free will enable scientists to monitor microglia activities in nervous system. Microglia is activated in response to a number of different pathological states within the CNS including injury, ischemia and infection. Microglial cell density is region specific and they comprise 5-20% (region dependant) of all cell in human brain. In order to properly understand their role in the context of CNS injury and diseases, it is vital that we are able to properly identify microglia and characterize between the activation state.
In this project, we develop Fluorescence lifetime Imaging Microscopy based technique to characterize microglia functional state. Further we also develop neural netowrk based machine learning tool to visualize microglia in vivo using FLIM data.
Spectra lifetime acquisition
The excited state lifetime of a fluorophore together with its fluorescence emission spectrum provide information that can yield valuable insights into the nature of a fluorophore and its microenvironment. However, it is difficult to obtain both channels of information in a conventional scheme as detectors are typically configured either for spectral or lifetime detection. We present a fiber-based method to obtain spectral information from a multiphoton fluorescence lifetime imaging (FLIM) system. This is made possible using the time delay introduced in the fluorescence emission path by a dispersive optical fiber coupled to a detector operating in time-correlated single-photon counting mode. This add-on spectral implementation requires only a few simple modifications to any existing FLIM system and is considerably more cost-efficient compared to currently available spectral detectors.
Paper link: http://dx.doi.org/10.1117/1.JBO.25.1.014506
Other collabarative research work
FLIM development (acquisition and analysis)
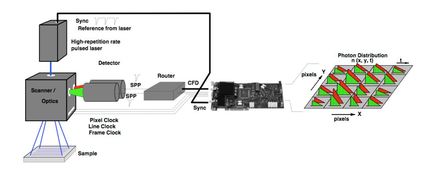
I work on the OWS (optical workstation) and SLIM (Spectral Lifetime Imaging Microscope) in our lab for investigating new methods and to acquire data as well as I manage FLIM collaborations across campus on these scopes. Both of these are laser scanning multiphoton scopes and are home-built. They are inverted scope and use a 80MHz Ti:sa laser for excitation.
I work on the software development for acquiring FLIM data as well as work on software development to analyze them. I look into FLIM data to understand metabolic changes in cell and detect cancer. FLIM is a promising tool for cancer detection specially breast cancer and there are many published material on this.
I am also investigating ways to extract spectral information from FLIM images. This will add one additional dimension of information to the already existing information layer. Combined Spectral and Lifetime Imaging(SLIM) will enable researchers to look at multiple fluorophores having different spectra combined with their fluorescence and lifetime measurement.
Despite its benefits, FLIM is slow compared to other laser scanning microscopy technique. That is a major limitation for studying various disease using FLIM or clinical implementation.
Right now I am working on overcoming limitations to acquire and analyse FLIM images faster. I am working with newer SPC-150 board with FIFO acquisition to acquire image faster with modification of software architecture to run live analysis during acquisition.
SLIM Curve
On the analysis front, we are have developed an ImageJ plugin(http://imagej.net/SLIM_Curve) which uses SLIM Curve exponential curve fitting library to analyze FLIM data. The plugin is written in Java with the core curve fitting routine written in C.
Other Analysis tool
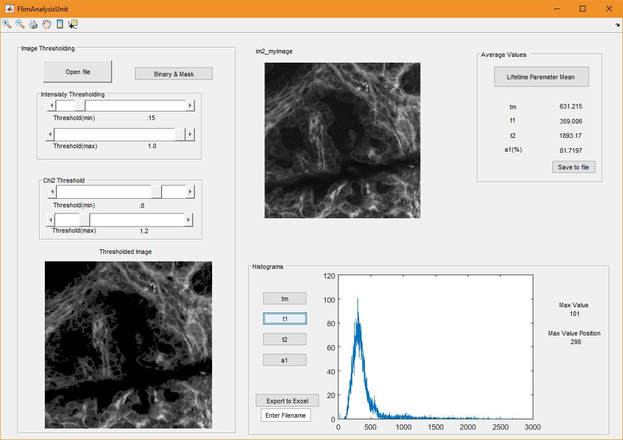
I have also developed small projects to analyze FLIM data more intuitively and easily. For example, this GUI based tool is a post-processing tool to analyze FLIM data exported from SPCImage software
WiscScan
Apart from lifetime imaging I am also a developer of WiscScan which is the primary software for controlling all the microscopes . It controls all the hardware component and performing scanning. WiscScan combines nearly all the functionality of a laser-scanning microscope/microbeam apparatus into a single software package. Our team regularly add new hardware feature and software feature in it. The goal of WiscScan is to automate all the processes related with acquiring image in microscope.
Google scholar link
Undergraduate Research
Breast Cancer Detection
My Undergrad thesis is "UWB microwave imaging via modified beamforming for early detection of breast cancer and FPGA implementation of the signal processing algorithm". This work was supervised by Dr A.B.M. Harun-ur Rashid. Here we proposed a novel beamforming algorithm for locating location of tumor. This method exploits the dielectric contrast of the normal tissue and malignant tissue. We use a crossed bow-tie antenna as transmitter and receiver for sending and receiving microwave signals. A 3-D FIT (Finite Integration Technique) model is used as breast model. The antenna is positioned in various location and the received signal was passed through the beamformer and image is formed. The beamformer is designed with adaptive weighting to compensate both propagation attenuation and lossy medium effect. Inspite of using using the traditional delay-and-sum approach, new delay-and-product technique is used in beamforming. This modified beamforming approach is shown to outperform its previous counterparts in terms of resolution and sensitivity. The simulation was done in CST. The whole signal processing algorithm was implemented in FPGA board. We used Altera DE2 board using cyclone 2 FPGA. The dissertation can be found here. The algorithm was published in ICECE 2010.
Fault detection and localization of CMOS circuits
Alongside my undergrad thesis, I also worked on fault detection and localization of CMOS circuits. We worked on this topic under supervision of Dr A.B.M Harun ur Rashid, Hamidur Rahman and Upal Mahbub. Here we developed an automated method based on wavelet analysis of IDD current to detect and localize faults in CMOS chips. The IDD current and its wavelet transform of the circuit under test are compared with those of a reference faultless circuit for detection and localization of fault. Here a novel similarity index(SI) was proposed for detection and localization of fault. The whole system was implemented in FPGA. Here we used Altera DE2 board with cyclone 2 FPGA. The findings are submitted to IET circuits, devices and systems for review.
Loss less image codec
Apart from this major reseach works, in my sophomore year in university I developed a lossless image codec in MATLAB. This was developed as a term project.